Our Research
Ortholist 2: genome-wide homology between
humans and C.elegans
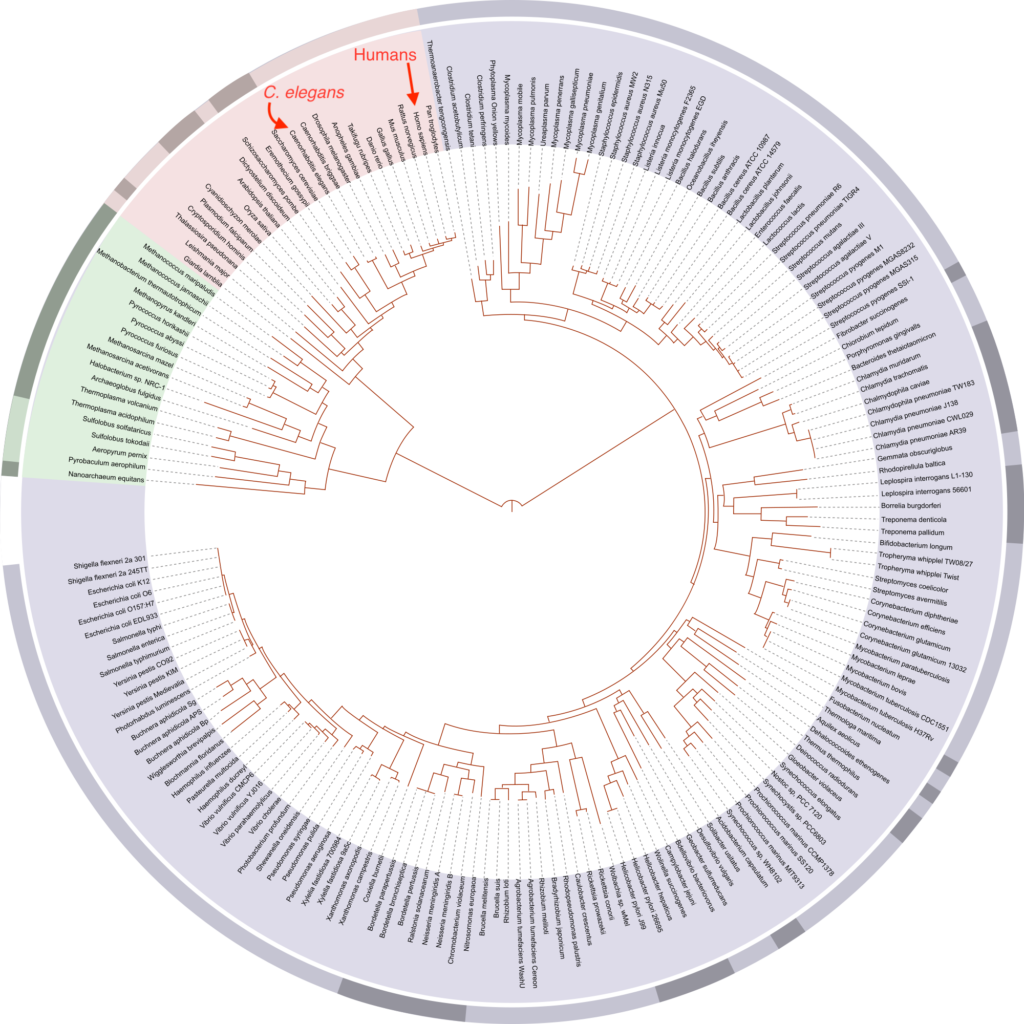
To focus our C. elegans studies on the genes most likely to be relevant to human biology and disease we (in conjunction with the Greenwald Lab) undertook a genome-wide approach to identify all the evolutionarily related genes (orthologs and paralogs) between worms and humans. We first published a compendium of human-C .elegans homologs in 2011. We have now re-visited and updated this study to assess how changes in genome annotations, updates to orthology predictions methods, and the inclusion of new predictions methods affects the landscape of predicted C. elegans-human homologs. The resulting updated database can be queried via an online tool found here.
Relevant literature:
Relevant literature:
- Shaye and Greenwald. PLoS ONE, 2011.
- Kim et al. Genetics, 2018.
The ExCa as a model to study FSGS-associated genes
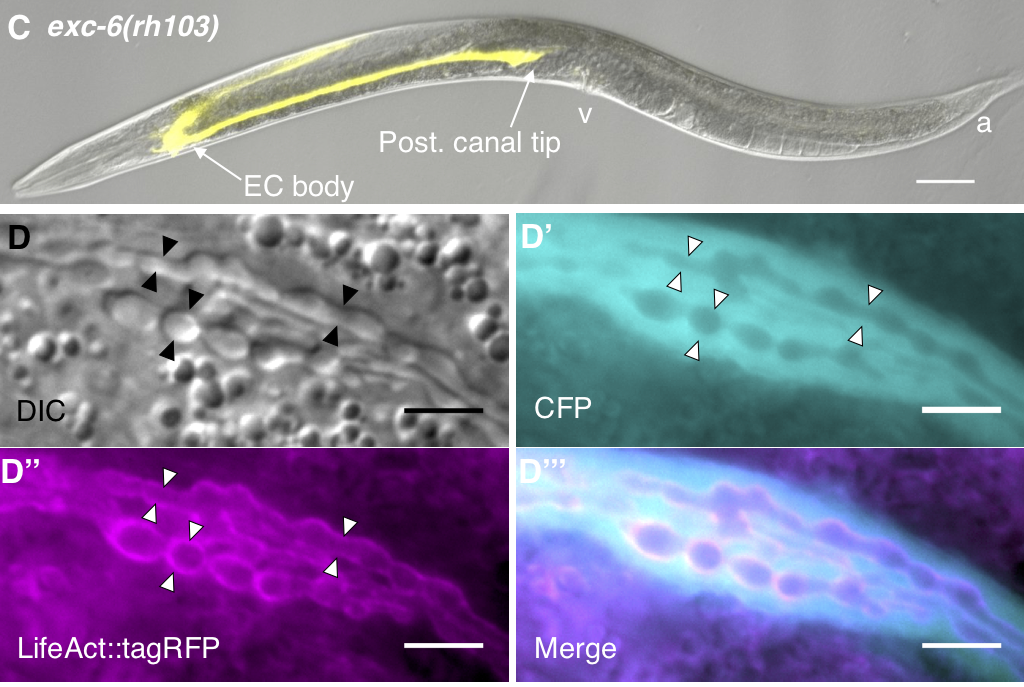
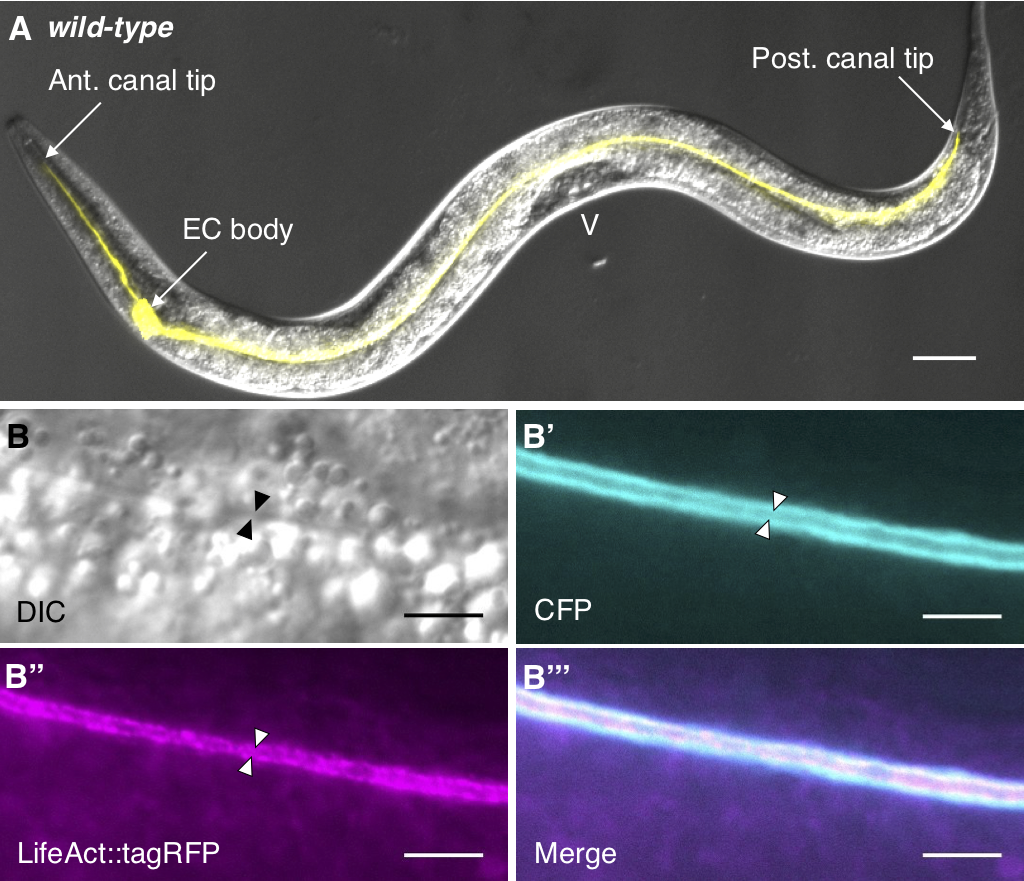
The C. elegans excretory canal (ExCa) cell provides a model to identify and study genes involved in tubulogenesis. This cell collects and excretes excessive fluids, which is critical for homeostasis and osmoregulation.
Genes that regulate development and maintenance of the ExCa have homologs in humans that have been implicated in kidney and vascular disease.
We cloned exc-6, a gene involved in ExCa development and found that it is an ortholog of the human gene INF2, which encodes a Formin that when mutated causes the kidney disease Focal Segmental Glomerulosclerosis FSGS.
Our work demonstrated that EXC-6 and INF2 function is conserved, determined the effect of disease-causing mutations and demonstrated a role for EXC-6/INF2 in regulating the microtubule cytoskeleton.
We have also studied a second worm INF2 ortholog, called inft-2, which we found is important for F-actin accumulation in the ExCa and is regulated by a third member of the formin family in C. elegans called cyk-1, the ortholog of vertebrate DIAPH.
We are currently using the ExCa to further analyze the role and regulation of INF2, as well as to study other conserved FSGS-associated genes, to better understand disease mechanisms with the goal of uncovering possible therapeutic targets.
Relevant literature:
Relevant literature:
- Shaye and Greenwald. Developmental Cell, 2015.Shaye and Greenwald. Development, 2016.
Discovery and analysis of angiogenesis regulators
Defects in angiogenesis (the formation of new blood vessels from pre-existing ones) can lead to congenital vascular disease, while tumor-induced angiogenesis promotes cancer survival and metastasis.
Tubulogenesis is a key biological process required for angiogenesis, and several genes involved in angiogenesis are also involved in ExCa tubulogenesis (e.g., the chloride intracellular channel EXC-4, and the kinases WNK1, SPAK and OXSR1).
We are combining the power of C. elegans genetics with well-established angiogenic assays to discover and analyze conserved regulators of tubulogenesis in general, and angiogenesis in particular.